Bloom Filters: A Powerful Tool
(credit: image source)
BloomFilter is a new Ruby library (available as a gem - gem install bloomfilter) by Bryan Duxbury that provides operations to create and query "bloom filters", an extremely space-efficient "probablistic data structure" that makes it quick and easy to test set membership. This all sounds incredibly geeky and uninteresting until you discover how bloom filters can be used to make things like ultra-fast, low-overhead spell checkers, spam filters, stop word removers, and other tools that require checking two sets of data against each other. Bryan's use for bloom filters (and BloomFilter) was to search a big list of 500 million hashes against a set of 40 million hashes. Given the size of each hash, coupled with the large quantity, a major, memory hungry task was looming. Bloom filters enabled the task to reduce from a 20 day running time to just 8 hours.
BloomFilter is partially influenced and based on a library I wrote a couple of months ago called BloominSimple (not available as a gem, but the code is simple). In my case I wrote BloominSimple to perform super-fast stop word removal (removing short, meaningless words from content). Using a "traditional" array based technique proved too slow on large collections of text, but with a bloom filter solution the time was reduced by an order of magnitude. Another library called sBloomFilter that uses C is also available, although I'm unfamiliar with its feature set.
Bloom filters are useful to have in your programming arsenal, so if you're not already familiar with them, there might be a way to significantly improve performance on some of your heavier comparison routines.




September 13, 2007 at 12:32 am
It sounds awesome. Maybe I will try when time's right.
September 24, 2007 at 11:42 am
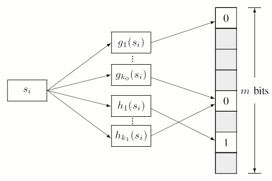
It is a very simple algorithm. In the image, s is the string, g's are some hashing functions whose output is a bit position in the array.
insertion -
s is passed to all the g's and their output is marked as 1 in the array
searching
s is passed to all the g's and their output position is queried. If all the positions have 1, then the string was inserted once, and is a positive.
Disadvantage is false positives, if many strings are inserted, some of combined the 1's may represent a string not yet inserted.